I wrote before about my current work on identifier mapping. Briefly, each of the many different databases for genes and metabolites uses its own system of identifiers. This creates big headaches when you want to compare things from different databases. You’ll have to do some work to correlate them, which is what we call the identifier mapping problem.
Why does this problem exist in the first place? Wouldn’t it be really fantastic if everybody would always use the same identifiers everywhere? I don’t think that’s ever going to happen. There are practical reasons for that, but there are also fundamental problems that can never be solved.
Scientific databases are organized in a way that reflects the mindset of the scientists that created them. I noticed the same argument in an essay by Clay Shirky about the semantic web:
Because meta-data describes a worldview, incompatibility is an inevitable by-product of vigorous argument. It would be relatively easy, for example, to encode a description of genes in XML, but it would be impossible to get a universal standard for such a description, because biologists are still arguing about what a gene actually is. There are several competing standards for describing genetic information, and the semantic divergence is an artifact of a real conversation among biologists. You can’t get a standard til you have an agreement, and you can’t force an agreement to exist where none actually does.
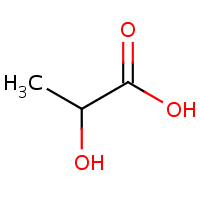
Lactic Acid
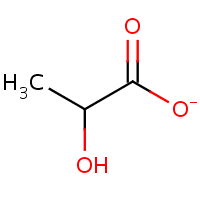
Lactate
Here is another example from the context of bioinformatics: a chemist might create separate identifiers for lactate and lactic acid. To a chemist, these are two different things, lactate is missing a hydrogen atom and it’s even negatively charged. But when dissolved in water these two rapidly convert into each other, making them practically indistinguishable. So a chemistry oriented database such as ChEBI describes them separately (CHEBI:24996 and CHEBI:28358) whereas a biological database such as HMDB puts both in a single record (HMDB00190) World views have affected the way these databases are set up.
By the way, the article quoted above is also an argument against the whole idea of the Semantic Web of Life Sciences (SWLS), but that’s subject matter for another post.
Tags: bridgedb, identifier mapping problem, identifiers, semantic web
The easiest way is Standardizer (ChemAxon) also the use of InChI can be helpful but with some limitatons…
Yes, there are many possible workarounds. However, and this is the point of the post, you’ll never be able to fix the source of the problem.
[…] your hearts content. BridgeDb is all about identifier mapping, which I blogged about before (here, here and […]